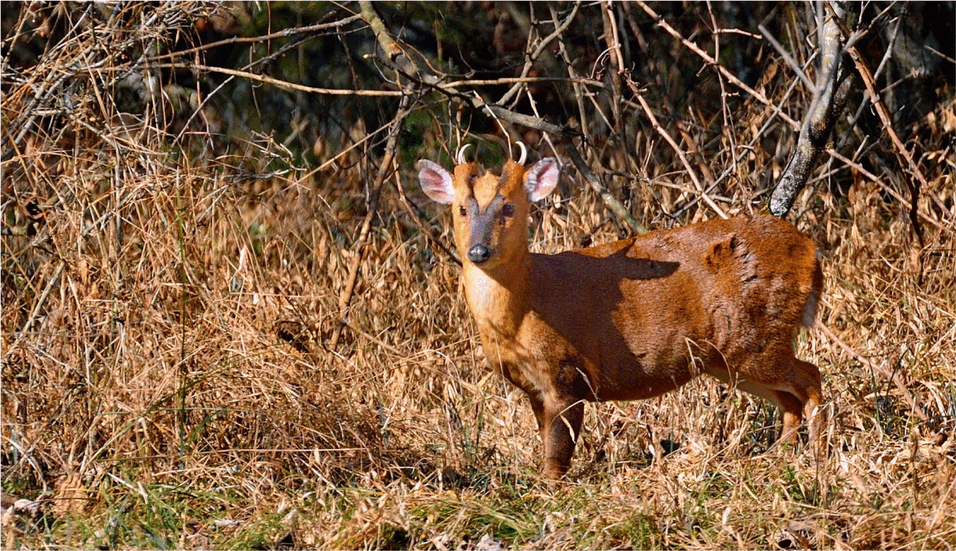
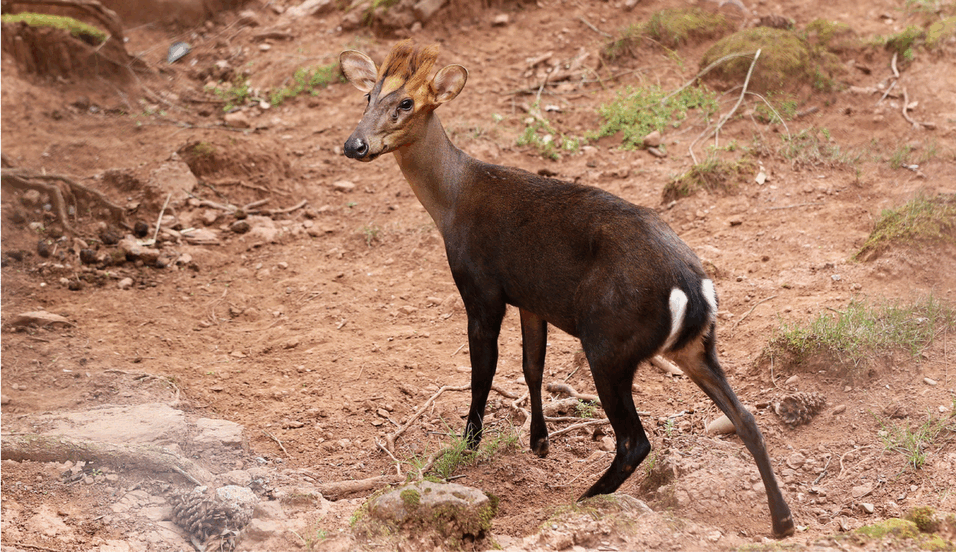
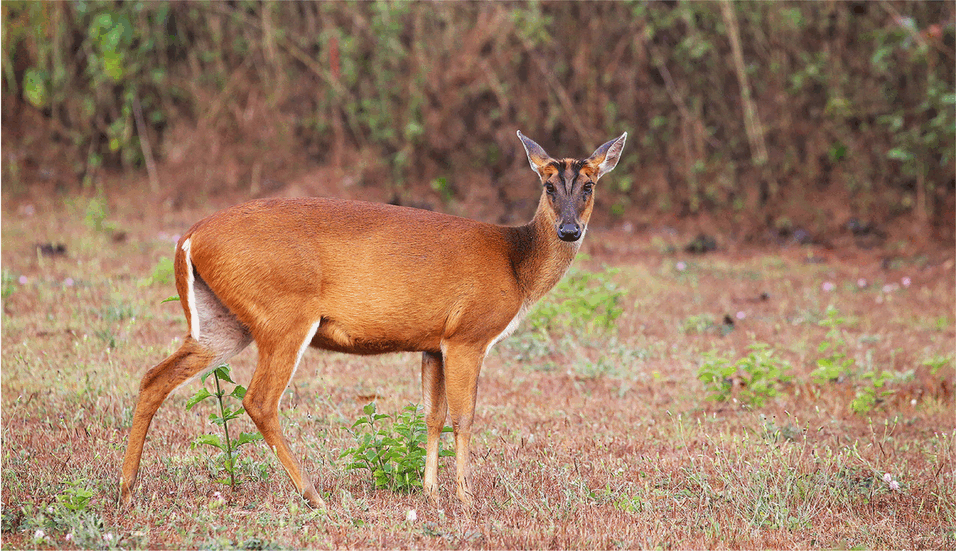
Muntjacs, also known as ‘barking deer’ for their distinctive call, show the greatest diversity of chromosome numbers among mammals. They range from 46 in Chinese muntjac to 6 or 7 chromosomes in Indian muntjac during the course of only 3 million years of their evolution. In an article in Nature Communications, scientists from the University of Vienna, the Chinese Academy of Sciences and the Northwestern Polytechnical University deciphered the genomes of these curious deer species and examined the causes of such dramatic chromosome changes, and the consequences of chromosome reduction for the overall genome.
Exotic pets for studying dramatic chromosome evolution
In the worst case, dramatic chromosome rearrangements can lead to cancer, but a group of deer species called muntjacs have coped with them extremely well during their evolution. Most muntjac species are mainly distributed in Asia. Some species like black muntjacs have become endangered due to recent disruption of habitat; while others like Chinese muntjacs, also called ‘little muntjac’ in Chinese, were introduced to the UK and become widespread in rural areas. Because of their small body size, tamed character and the barking sound that they make, some muntjacs are even kept as pets. But scientists have been curious about these lesser-known deer for another reason. Indian muntjacs only have 6 chromosomes in females, and 7 chromosomes (the additional chromosome is the Y chromosome) in males, making them the species with the lowest number of chromosomes known in mammals. What is even more astonishing is that their closely related species, the Chinese muntjacs, have the same chromosome number as human, i.e., 46, even though the two species only diverged within 3 million years.
Massive chromosome fusions, few genomic changes
the intriguing case of muntjacs was already noticed in the 1980 by the Nobel Laureate Barbara McClintock. She described the radical change in chromosome number in a lecture as a case of ‘genomic shocking’ event. But so far, neither the causes for the massive disruption nor its consequences were known. Recently, groups of Qi Zhou from University of Vienna and Zhejiang University of China, Wen Wang from Northwestern Polytechnical University of China, and Fuwei Wen at Chinese Academy of Sciences, collaborated and produced high-quality chromosomal genomes of several muntjac and some other related deer species. From the new genomic data, they inferred that about 1 million years ago a glaciation event in Southwest China led to the drastic population decline in those muntjac species that nowadays have much less chromosomes. Chinese muntjacs, that have the same chromosome number as humans, did not experience the decline as they were likely distributed in another area which was not affected by the steep decline in temperature. The historical population shrinkage might be associated with the observed massive chromosome fusions in muntjacs. Another possible contributing factor may be located within the muntjacs’ genome. There are complex repeat sequences, located at the chromosome ends that are specific to the muntjacs. These repeat sequences are on the one hand prone to sequence breaks and on the other hand, they have a high chance to ‘connect’ to each other due to their sequence similarities. Both factors may contribute to chromosome fusions.
Zhou and his colleagues also found, that the chromosome fusions neither influence the genomic sequences, nor how the genome is folded in its three-dimensional structures, meaning that for the assembly of the genome or for the content of the genes themselves it does not seem to matter much if the chromosomes are separated or fused, at least for the case of muntjacs. A comparison between Indian muntjacs and Chinese muntjacs found that their overall genome architectures are similar to each other, despite their great differences in chromosome numbers. This may explain why even hybrids between the two species can exist like one individual in a zoo in Shanghai in the 1970. An exception of this can be found in the black muntjac, which displays a male-specific chromosome inversion (so it is evolving like a Y chromosome without recombination). In this inverted genomic region, both the sequences and genome folding have greatly changed relative to its counterpart chromosome without the inversion. Overall, this study indicates, that the type (fusion vs. inversion) rather than the number of chromosome rearrangements matter a lot to the genome function.
Publication in Nature Communications:
Yin, Y., Fan, H., Zhou, B. et al. Molecular mechanisms and topological consequences of drastic chromosomal rearrangements of muntjac deer. Nat Commun 12, 6858 (2021). doi.org/10.1038/s41467-021-27091-0